Plots a Venn diagram showing overlapping significant proteins
Source:R/plots.R
plot_venn_diagram.RdPlots a Venn diagram showing overlapping significant proteins
Usage
plot_venn_diagram(
res,
...,
comparisons = "all",
colors = c("#b3e2cd", "#fdcdac", "#cbd5e8", "#f4cae4", "#e6f5c9"),
names = NULL
)Arguments
- res
A data frame with results from get_DEPresults()
- ...
Additional results data frames (optional)
- comparisons
A character vector specifying which comparisons to include. The default 'all' considers all comparisons present in res.
- colors
The colors used for the Venn diagram.
- names
(optional). Specifies the labels for the different subsets. Defaults to the comparison names in colnames(res)
Examples
se <- prepare_se(report.pg_matrix, expDesign)
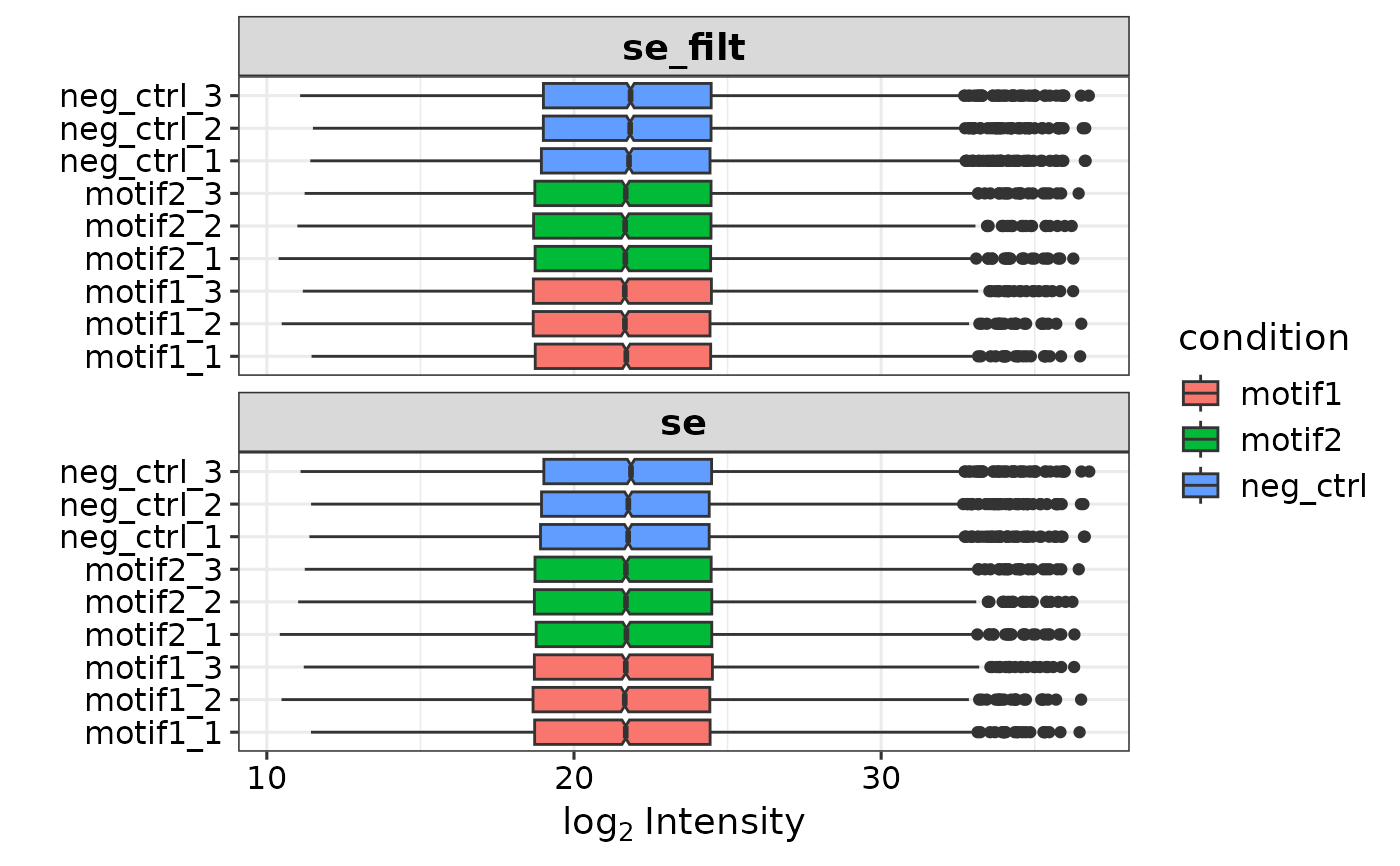 #> Imputing along margin 2 (samples/columns).
#> [1] 0.3066195
#> Imputing along margin 1 (features/rows).
#> Warning: 36 rows with more than 50 % entries missing;
#> mean imputation used for these rows
#> Cluster size 5574 broken into 3662 1912
#> Cluster size 3662 broken into 1462 2200
#> Done cluster 1462
#> Cluster size 2200 broken into 1110 1090
#> Done cluster 1110
#> Done cluster 1090
#> Done cluster 2200
#> Done cluster 3662
#> Cluster size 1912 broken into 1379 533
#> Done cluster 1379
#> Done cluster 533
#> Done cluster 1912
res <- get_DEPresults(se, type = 'all')
#> Tested contrasts: neg_ctrl_vs_motif1, neg_ctrl_vs_motif2, motif1_vs_motif2
plot_venn_diagram(res) # Default option
#> INFO [2025-10-14 08:00:14] [[1]]
#> INFO [2025-10-14 08:00:14] venn_list
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $filename
#> INFO [2025-10-14 08:00:14] NULL
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $disable.logging
#> INFO [2025-10-14 08:00:14] T
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fill
#> INFO [2025-10-14 08:00:14] colors
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontface
#> INFO [2025-10-14 08:00:14] [1] "bold"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $cat.fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $lty
#> INFO [2025-10-14 08:00:14] [1] 0
#> INFO [2025-10-14 08:00:14]
#> Imputing along margin 2 (samples/columns).
#> [1] 0.3066195
#> Imputing along margin 1 (features/rows).
#> Warning: 36 rows with more than 50 % entries missing;
#> mean imputation used for these rows
#> Cluster size 5574 broken into 3662 1912
#> Cluster size 3662 broken into 1462 2200
#> Done cluster 1462
#> Cluster size 2200 broken into 1110 1090
#> Done cluster 1110
#> Done cluster 1090
#> Done cluster 2200
#> Done cluster 3662
#> Cluster size 1912 broken into 1379 533
#> Done cluster 1379
#> Done cluster 533
#> Done cluster 1912
res <- get_DEPresults(se, type = 'all')
#> Tested contrasts: neg_ctrl_vs_motif1, neg_ctrl_vs_motif2, motif1_vs_motif2
plot_venn_diagram(res) # Default option
#> INFO [2025-10-14 08:00:14] [[1]]
#> INFO [2025-10-14 08:00:14] venn_list
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $filename
#> INFO [2025-10-14 08:00:14] NULL
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $disable.logging
#> INFO [2025-10-14 08:00:14] T
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fill
#> INFO [2025-10-14 08:00:14] colors
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontface
#> INFO [2025-10-14 08:00:14] [1] "bold"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $cat.fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $lty
#> INFO [2025-10-14 08:00:14] [1] 0
#> INFO [2025-10-14 08:00:14]
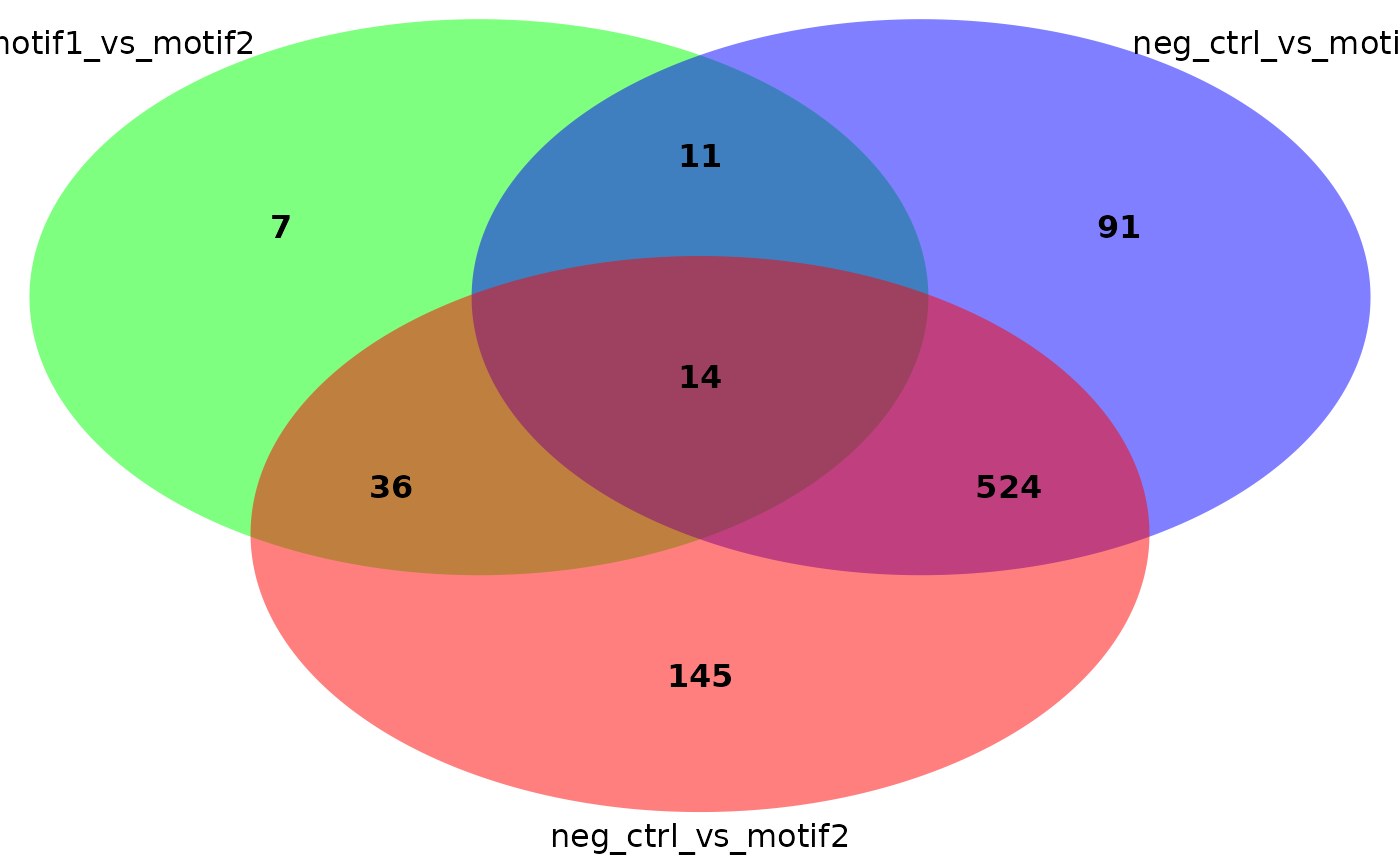
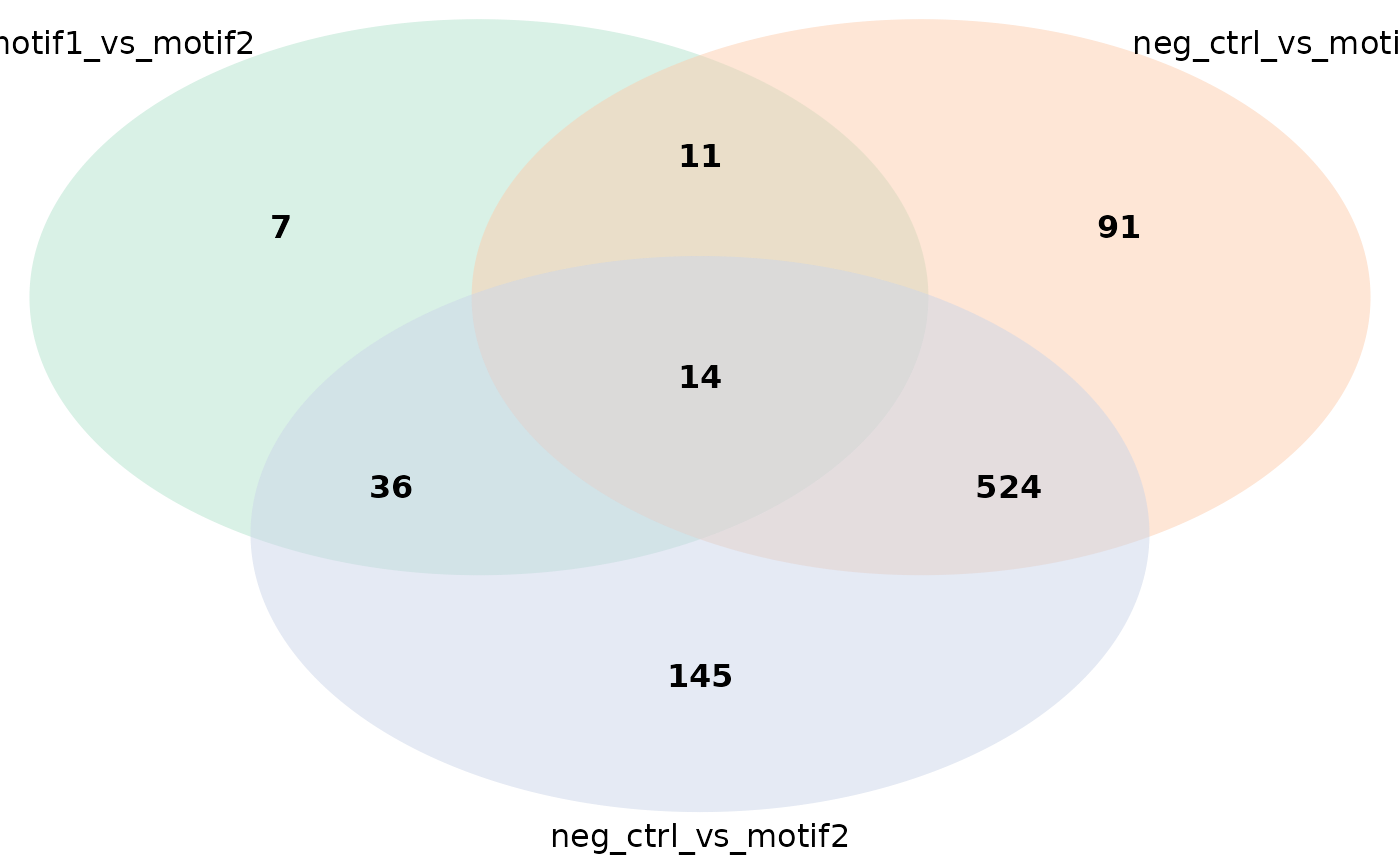 plot_venn_diagram(res, colors = c('green', 'blue', 'red')) # Uses non-default
#> INFO [2025-10-14 08:00:14] [[1]]
#> INFO [2025-10-14 08:00:14] venn_list
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $filename
#> INFO [2025-10-14 08:00:14] NULL
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $disable.logging
#> INFO [2025-10-14 08:00:14] T
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fill
#> INFO [2025-10-14 08:00:14] colors
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontface
#> INFO [2025-10-14 08:00:14] [1] "bold"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $cat.fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $lty
#> INFO [2025-10-14 08:00:14] [1] 0
#> INFO [2025-10-14 08:00:14]
plot_venn_diagram(res, colors = c('green', 'blue', 'red')) # Uses non-default
#> INFO [2025-10-14 08:00:14] [[1]]
#> INFO [2025-10-14 08:00:14] venn_list
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $filename
#> INFO [2025-10-14 08:00:14] NULL
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $disable.logging
#> INFO [2025-10-14 08:00:14] T
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fill
#> INFO [2025-10-14 08:00:14] colors
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontface
#> INFO [2025-10-14 08:00:14] [1] "bold"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $cat.fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $lty
#> INFO [2025-10-14 08:00:14] [1] 0
#> INFO [2025-10-14 08:00:14]
 # colors
plot_venn_diagram(res, comparisons = c('neg_ctrl_vs_motif1',
'neg_ctrl_vs_motif2')) # Only
#> INFO [2025-10-14 08:00:14] [[1]]
#> INFO [2025-10-14 08:00:14] venn_list
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $filename
#> INFO [2025-10-14 08:00:14] NULL
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $disable.logging
#> INFO [2025-10-14 08:00:14] T
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fill
#> INFO [2025-10-14 08:00:14] colors
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontface
#> INFO [2025-10-14 08:00:14] [1] "bold"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $cat.fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $lty
#> INFO [2025-10-14 08:00:14] [1] 0
#> INFO [2025-10-14 08:00:14]
# colors
plot_venn_diagram(res, comparisons = c('neg_ctrl_vs_motif1',
'neg_ctrl_vs_motif2')) # Only
#> INFO [2025-10-14 08:00:14] [[1]]
#> INFO [2025-10-14 08:00:14] venn_list
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $filename
#> INFO [2025-10-14 08:00:14] NULL
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $disable.logging
#> INFO [2025-10-14 08:00:14] T
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fill
#> INFO [2025-10-14 08:00:14] colors
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $fontface
#> INFO [2025-10-14 08:00:14] [1] "bold"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $cat.fontfamily
#> INFO [2025-10-14 08:00:14] [1] "sans"
#> INFO [2025-10-14 08:00:14]
#> INFO [2025-10-14 08:00:14] $lty
#> INFO [2025-10-14 08:00:14] [1] 0
#> INFO [2025-10-14 08:00:14]
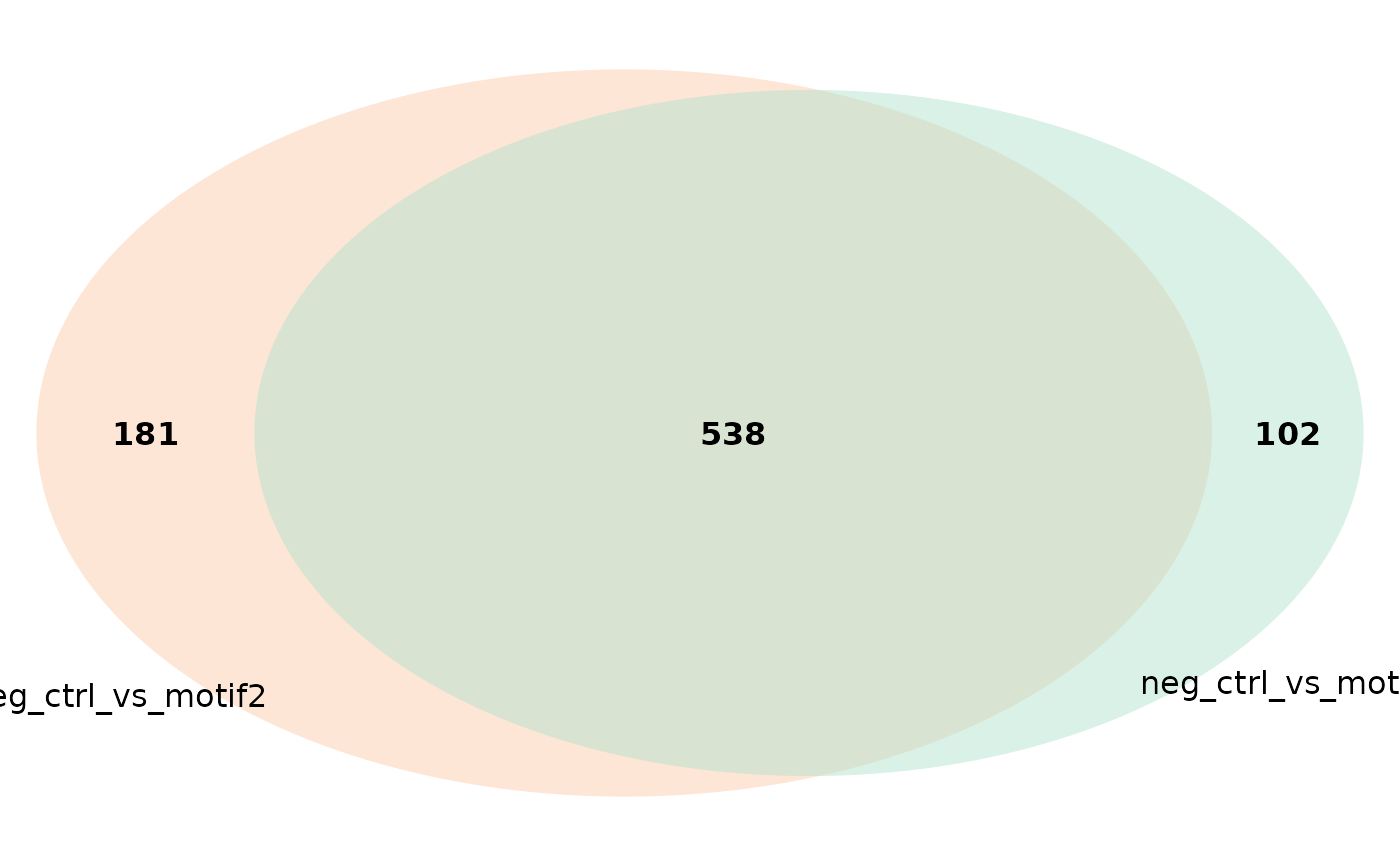 # includes two comparisons
# includes two comparisons